Note
Click here to download the full example code
Generating Cluster Graphs
This example shows how to find the communities in a graph, then contract each community into a single node using igraph.clustering.VertexClustering. For this tutorial, we’ll use the Donald Knuth’s Les Miserables Network, which shows the coapperances of characters in the novel Les Miserables.
import igraph as ig
import matplotlib.pyplot as plt
We begin by load the graph from file. The file containing this network can be downloaded here.
g = ig.load("./lesmis/lesmis.gml")
Now that we have a graph in memory, we can generate communities using
igraph.Graph.community_edge_betweenness() to separate out vertices into
clusters. (For a more focused tutorial on just visualising communities, check
out Communities).
communities = g.community_edge_betweenness()
For plots, it is convenient to convert the communities into a VertexClustering:
communities = communities.as_clustering()
We can also easily print out who belongs to each community:
for i, community in enumerate(communities):
print(f"Community {i}:")
for v in community:
print(f"\t{g.vs[v]['label']}")
Community 0:
Myriel
Napoleon
MlleBaptistine
MmeMagloire
CountessDeLo
Geborand
Champtercier
Cravatte
Count
OldMan
Community 1:
Labarre
Valjean
MmeDeR
Isabeau
Gervais
Bamatabois
Simplice
Scaufflaire
Woman1
Judge
Champmathieu
Brevet
Chenildieu
Cochepaille
Community 2:
Marguerite
Tholomyes
Listolier
Fameuil
Blacheville
Favourite
Dahlia
Zephine
Fantine
Perpetue
Community 3:
MmeThenardier
Thenardier
Javert
Pontmercy
Eponine
Anzelma
Gueulemer
Babet
Claquesous
Montparnasse
Brujon
Community 4:
Cosette
Woman2
Gillenormand
Magnon
MlleGillenormand
MmePontmercy
MlleVaubois
LtGillenormand
BaronessT
Toussaint
Community 5:
Fauchelevent
MotherInnocent
Gribier
Community 6:
Boulatruelle
Community 7:
Jondrette
MmeBurgon
Community 8:
Gavroche
Marius
Mabeuf
Enjolras
Combeferre
Prouvaire
Feuilly
Courfeyrac
Bahorel
Bossuet
Joly
Grantaire
MmeHucheloup
Community 9:
MotherPlutarch
Community 10:
Child1
Child2
Finally we can proceed to plotting the graph. In order to make each community stand out, we set “community colors” using an igraph palette:
num_communities = len(communities)
palette1 = ig.RainbowPalette(n=num_communities)
for i, community in enumerate(communities):
g.vs[community]["color"] = i
community_edges = g.es.select(_within=community)
community_edges["color"] = i
We can use a dirty hack to move the labels below the vertices ;-)
g.vs["label"] = ["\n\n" + label for label in g.vs["label"]]
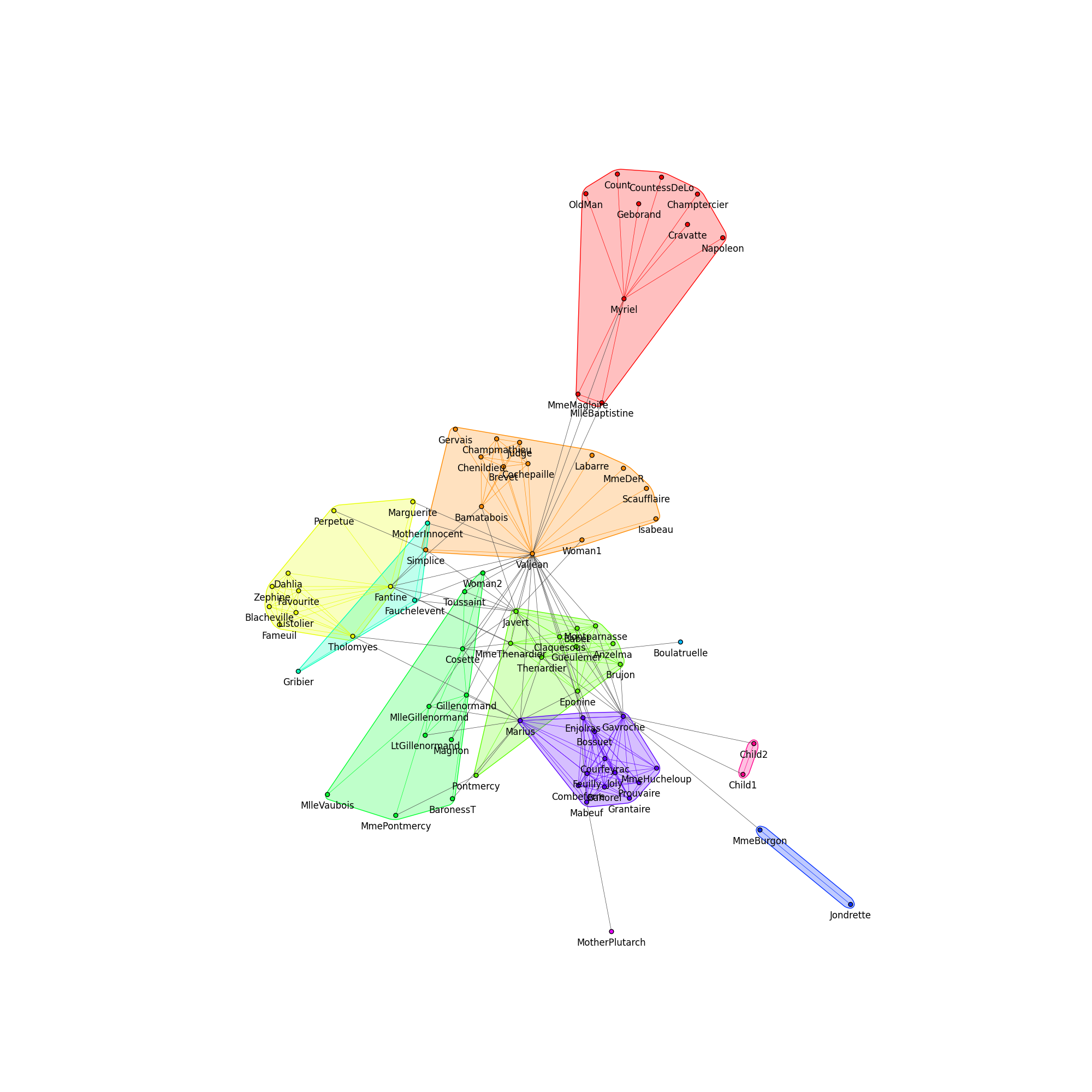
Finally, we can plot the communities:
fig1, ax1 = plt.subplots()
ig.plot(
communities,
target=ax1,
mark_groups=True,
palette=palette1,
vertex_size=0.1,
edge_width=0.5,
)
fig1.set_size_inches(20, 20)
Now let’s try and contract the information down, using only a single vertex to represent each community. We start by defining x, y, and size attributes for each node in the original graph:
layout = g.layout_kamada_kawai()
g.vs["x"], g.vs["y"] = list(zip(*layout))
g.vs["size"] = 1
g.es["size"] = 1
Then we can generate the cluster graph that compresses each community into a
single, “composite” vertex using
igraph.VertexClustering.cluster_graph():
cluster_graph = communities.cluster_graph(
combine_vertices={
"x": "mean",
"y": "mean",
"color": "first",
"size": "sum",
},
combine_edges={
"size": "sum",
},
)
Note
We took the mean of x and y values so that the nodes in the cluster graph are placed at the centroid of the original cluster.
Note
mean, first, and sum are all built-in collapsing functions,
along with prod, median, max, min, last, random.
You can also define your own custom collapsing functions, which take in a
list and return a single element representing the combined attribute
value. For more details on igraph contraction, see
igraph.GraphBase.contract_vertices().
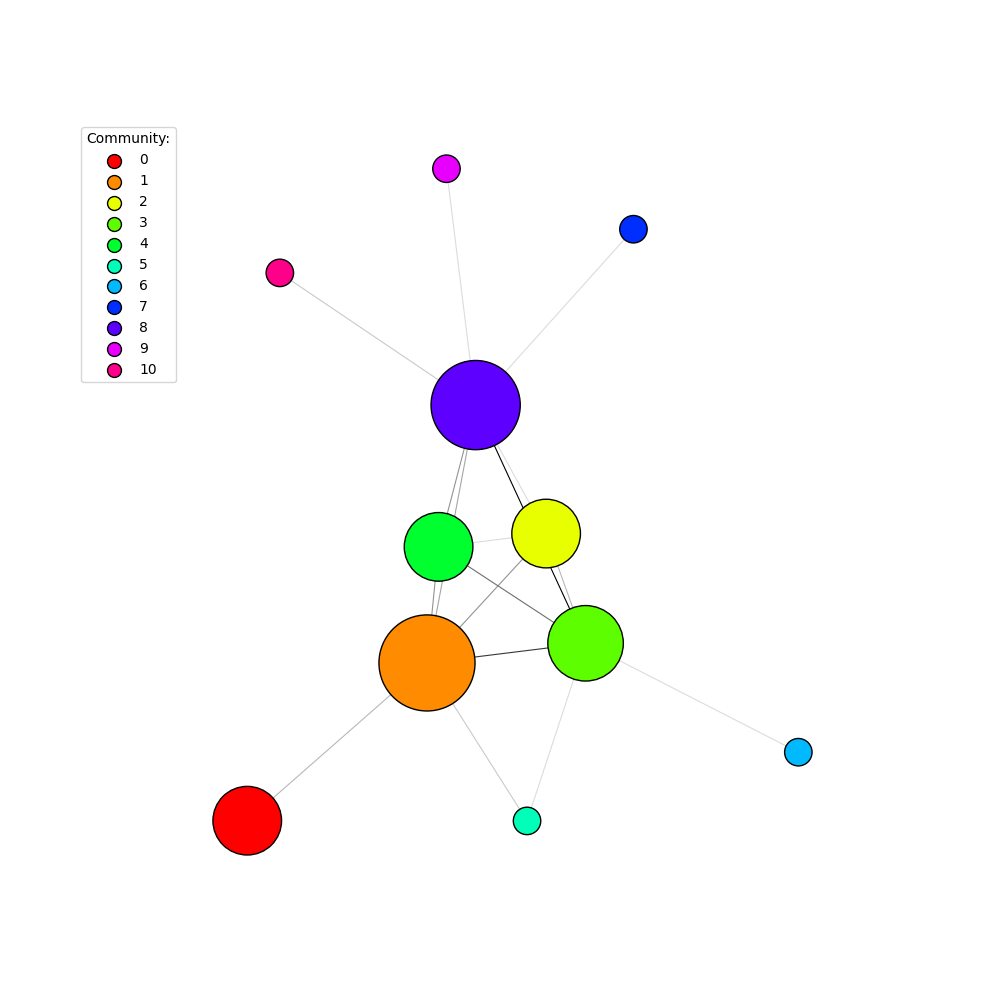
Finally, we can assign colors to the clusters and plot the cluster graph, including a legend to make things clear:
palette2 = ig.GradientPalette("gainsboro", "black")
g.es["color"] = [palette2.get(int(i)) for i in ig.rescale(cluster_graph.es["size"], (0, 255), clamp=True)]
fig2, ax2 = plt.subplots()
ig.plot(
cluster_graph,
target=ax2,
palette=palette1,
# set a minimum size on vertex_size, otherwise vertices are too small
vertex_size=[max(0.2, size / 20) for size in cluster_graph.vs["size"]],
edge_color=g.es["color"],
edge_width=0.8,
)
# Add a legend
legend_handles = []
for i in range(num_communities):
handle = ax2.scatter(
[], [],
s=100,
facecolor=palette1.get(i),
edgecolor="k",
label=i,
)
legend_handles.append(handle)
ax2.legend(
handles=legend_handles,
title='Community:',
bbox_to_anchor=(0, 1.0),
bbox_transform=ax2.transAxes,
)
fig2.set_size_inches(10, 10)
Total running time of the script: ( 0 minutes 1.052 seconds)